Table of Contents
DNA fingerprinting Class 12, also known as DNA typing or DNA profiling, is a powerful technique in genetics used to identify individuals based on their unique DNA patterns. Sir Alec Jeffreys was a geneticist at the University of Leicester in the UK studying the myoglobin gene in seal meat. He discovered homologous short repeating sequences shared between seals and humans.
These sequences, akin to minisatellites, were found to be highly variable genetic markers. Jeffreys and his team developed a radioactive probe to bind to these repeating sequences, unveiling unique patterns for each individual—a DNA “fingerprint.” This innovation marked a significant milestone in the field of forensic science and genetics.
What is DNA Fingerprinting Class 12?
DNA fingerprinting, also known as DNA profiling or genetic fingerprinting, is a scientific technique used to identify individuals based on their unique genetic makeup. It analyzes specific regions of an individual’s DNA sequence that vary from person to person. This method is widely used in forensic science, paternity testing, criminal investigations, and genetic research.
DNA Polymorphism
DNA polymorphism forms the basis for DNA fingerprinting and genome mapping. DNA polymorphism refers to variations in the DNA sequence. These variations arise due to mutations, which can occur in an individual’s somatic cells or germ cells. Mutations in repetitive DNA allelic sequence variation, where multiple alleles exist at a single locus in a population with a frequency greater than 0.01, is a common form of DNA polymorphism.
These variations are more likely to occur in non-coding DNA sequences, as mutations in these regions do not immediately affect reproductive fitness. If a mutation impacts an individual’s ability to have offspring, it cannot be passed down several generations. DNA polymorphism is essential for genetic analysis and molecular studies, serving as genetic markers, and can play a significant role in evolutionary processes and speciation.
VNTRs
Variable Number Tandem Repeats (VNTRs) are specific genomic regions where short sequences of DNA are repeated in tandem, and these repeats often vary in length among individuals, serving as inherited alleles. They are a specific subset of minisatellites, which are characterized by repetitive DNA sequences that repeat in tandem. VNTRs typically have repeat sequences ranging from ten to one hundred nucleotides, and the number of repeats can vary among individuals.
This variability in repeat numbers makes VNTRs valuable in applications like DNA fingerprinting and forensics, as they create unique genetic profiles. By extracting and analyzing VNTRs, each individual’s unique genetic profile can be determined, making it highly improbable for unrelated individuals to share the same allelic pattern when multiple VNTR markers are considered.
Steps of DNA fingerprinting Class 12
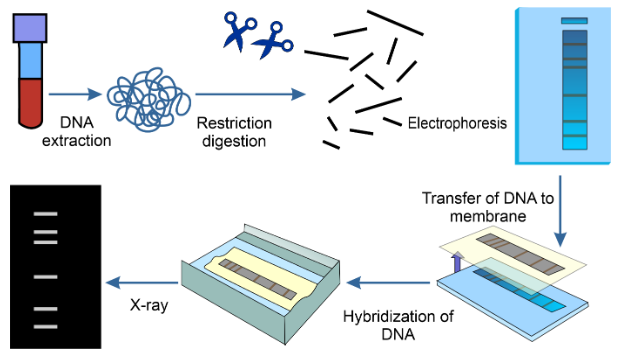
- Sample collection: The first step is to obtain a sample of cells containing DNA. This sample can come from various sources, such as skin cells, hair follicles, or blood.
- DNA extraction: DNA is extracted from the collected cells. This process involves breaking down the cell membranes and proteins to isolate the DNA molecules.
- Restriction enzyme digestion: In the original approach, known as restriction fragment length polymorphism (RFLP) analysis, the extracted DNA is cut at specific points along the DNA strands using proteins called restriction enzymes. These enzymes cleave the DNA at specific recognition sites.
- Gel electrophoresis: The resulting DNA fragments, which vary in length, are then separated by size using a technique called gel electrophoresis. The DNA fragments are loaded onto a gel and subjected to an electric current. Smaller fragments move faster through the gel, while larger fragments move more slowly.
- Blotting: After electrophoresis, the sorted DNA fragments are transferred onto a nylon membrane in a process called blotting. This step allows for further analysis of the DNA fragments.
- Probe hybridization: Synthetic DNA probes are used to identify specific regions of interest, such as minisatellites (variable number tandem repeats, VNTRs) or microsatellites (short tandem repeats, STRs). These probes are often labeled with radioactive markers or fluorescent tags. The probes will bind to complementary DNA sequences on the membrane.
- Autoradiography: In RFLP analysis, the membrane is exposed to X-ray film, and dark marks appear at the locations where the radioactive probes have bound to the DNA fragments. This creates a pattern of marks unique to each individual.
- Analysis: The resulting autoradiograph or fluorescent image is analyzed to compare the DNA patterns. The pattern of bands on the gel or membrane is like a fingerprint for the individual, and no two people (except identical twins) have the same pattern.
- Modern PCR-Based Approaches: While RFLP analysis was the original method for DNA fingerprinting Class 12, it has largely been replaced by polymerase chain reaction (PCR) and microsatellite or STR analysis. PCR allows for the amplification of specific DNA regions, and STRs are short repeat sequences that are highly variable among individuals. This approach is more sensitive, requires less DNA, and is better suited for forensic analysis.
Write the Applications for DNA Fingerprinting Class 12
DNA fingerprinting is a powerful technique used to identify individuals based on their unique genetic patterns. It has various applications across different fields, including forensic science, medicine, and agriculture.
-
Forensic Investigations:
- Helps in identifying criminals from biological evidence like blood, hair, or skin cells found at crime scenes.
- Used in solving cases related to murder, sexual assault, and theft.
-
Paternity and Maternity Testing:
- Determines biological relationships between parents and children.
- Useful in inheritance disputes and child custody cases.
-
Identification of Missing Persons and Disaster Victims:
- Helps in identifying unidentified bodies after natural disasters, accidents, or war casualties.
-
Medical and Genetic Research:
- Detects genetic disorders and hereditary diseases.
- Helps in organ transplantation by ensuring compatibility between donors and recipients.
-
Wildlife Conservation and Animal Breeding:
- Identifies endangered species and prevents poaching.
- Ensures the purity of breeding lines in livestock and pet industries.
-
Agriculture and Food Safety:
- Used in the identification of genetically modified (GM) crops.
- Helps in tracing food authenticity and preventing food fraud.
DNA fingerprinting Class 12 plays a crucial role in modern science and has significantly improved accuracy in criminal justice, medical diagnostics, and biological research.
Summary
DNA fingerprinting, a groundbreaking technique pioneered by Sir Alec Jeffreys, has evolved into a versatile tool with diverse applications. This method is rooted in the understanding of DNA polymorphism, variations in DNA sequences due to mutations, and relies on Variable Number Tandem Repeats (VNTRs), a subset of minisatellites. The steps of DNA fingerprinting Class 12 involve sample collection, DNA extraction, restriction enzyme digestion, gel electrophoresis, probe hybridisation, autoradiography, and analysis, with modern PCR-based approaches offering enhanced sensitivity.
DNA fingerprinting’s applications span forensic science, where it aids in solving crimes and establishing innocence; paternity and family relationship testing for legal and emotional purposes; genetic research and diversity studies; disaster victim identification; immigration and citizenship verification; tracing human migration and ancestry; wildlife conservation; and personalized medicine.
FAQs on DNA Fingerprinting
What is DNA fingerprinting?
DNA fingerprinting is a technique used to identify individuals based on unique patterns in their DNA. It relies on the analysis of DNA polymorphism, which leads to variations in DNA sequences.
How does DNA fingerprinting work?
DNA fingerprinting involves extracting DNA from a sample, cutting it using restriction enzymes, separating the fragments by gel electrophoresis, and then analyzing the pattern of DNA fragments, often using probes specific to Variable Number Tandem Repeats (VNTRs).
Who is the ‘father of DNA fingerprinting’?
Sir Alec Jeffreys, a geneticist at the University of Leicester in the UK.
Expand VNTR.
Variable number tandem repeats. These belong to a class of minisatellites.
How does DNA fingerprinting aid in paternity testing?
DNA fingerprinting allows for precise determination of paternity and family relationships by comparing DNA profiles. This has legal implications in child support cases and inheritance disputes.
Why is it called DNA fingerprinting?
The probability of having two people with the same DNA fingerprint that are not identical twins is very small. Each of us is genetically unique and this genetic variation could be used to identify individuals, as a conventional fingerprint does. Thus, it is called DNA fingerprinting.
When was DNA fingerprinting first used?
1986. DNA fingerprinting was first used in forensic science in 1986 when police in the UK requested Dr. Alec J. Jeffreys, of University of Leicester, to verify a suspect's confession that he was responsible for two rape-murders. Tests proved that the suspect had not committed the crimes.








